Feature
Physics is the new biology
They’ve done their chemistry and genetics. Now scientists are ready to explore the mechanics of being alive.
By Alexander GelfandAmy E. Shyer is watching a movie on her laptop.
It’s a stop-motion clip her lab has created of cells on the move. And it reveals a fundamental truth about why cells do what they do.
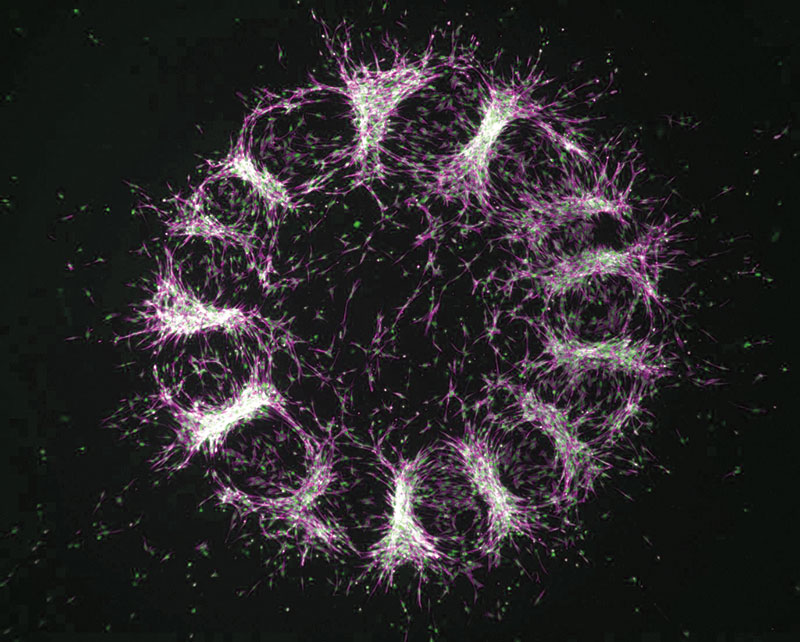
Shyer’s film shows what happens when skin cells, extracted from chicken embryos, are spread across a membrane that has just the right degree of rigidity. Frame by frame, the cells jostle and nudge one another until they begin to form small clumps. In living birds, these clumps would eventually become follicles, which would in turn sprout feathers.
Conventional wisdom says that the subtle dance these skin cells perform must be choreographed by genes buried deep within their nuclei. Those genes, so the story goes, express protein molecules that signal the cells to arrange themselves in patterns, migrating to form bits of tissue.
This model of morphogenesis, the process by which an organ takes shape, has been reinforced by decades of research that puts DNA at the center of all biological processes, from the healthy evolution of cells and tissues to the development of diseases like cancer and Alzheimer’s. As a result, says A. James Hudspeth, head of the Laboratory of Sensory Neuroscience, “biology has been a monoculture for the last two or three decades,” with scientists focused on using the tools of molecular biology to harvest the answers buried in our DNA.

That approach yields a theory of life that is simple and organized—DNA tells our cells what to do, and they do it—but that doesn’t tell the full story. A growing number of researchers are finding that cells are capable of responding to more than just their own DNA, and that genomics and biochemistry can’t explain everything.
Shyer and Hudspeth are interested in biomechanics: the same principles of force and motion, first established by Galileo and Newton, that allow engineers to build bridges and launch satellites. What they are finding is that the forces that act on cells, exerted by their neighbors and by the surfaces they live on—even the movements those cells make as they squirm, crawl, and otherwise go about their business—can be as essential to their functioning as genes and proteins, and may sometimes in fact trigger changes in gene expression and biochemistry.
The results can be surprising, even counterintuitive. Shyer’s research, for instance, indicates that the skin cells busily aggregating under her microscope are moving around not because of cues they receive from biochemical messaging, but only from the forces they exert as they push and pull themselves into formation.
“The cells are self-organizing,” explains Shyer, “and they’re doing it based on physical interactions.”
8,325The average number of feathers on a Plymouth Rock chicken.
More broadly, her findings suggest that purely mechanical processes—ones that emerge from the physical interactions of moving cells as they exert force and respond to it—are just as central to morphogenesis as genes and the biochemical signals they regulate. They may even, in some cases, trigger the genetic and biochemical processes that have occupied center stage for so long.
This mechanically oriented perspective is gaining currency across the biological sciences. Researchers are now exploring the biomechanics of phenomena ranging from hearing to DNA replication, often using technologies of their own invention. In so doing, they hope to illuminate the fundamental mechanisms that drive both normal and abnormal development, understand how diseases originate, and even create new opportunities for treatment and prevention.
Shyer, who is head of the Laboratory of Morphogenesis, first became interested in biomechanics as a graduate student at Harvard, where she worked to understand how the intestine develops its signature array of loops and coils.
According to the central dogma of her field, that configuration ought to have originated in a special pattern of gene expression—a chain reaction in which the activation of one gene after another produces a series of molecular events that mold the developing tissue. But try as they might, Shyer and her colleagues could not find such a pattern.
Instead, they discovered that the intestine’s distinctive shape emerged from what applied mathematicians and civil engineers call a “buckling problem”: the same phenomenon that causes the columns in a building to bend and warp under stress.
Mechanical events can sometimes drive molecular ones, and not the other way around.
For Shyer, the realization that an entirely mechanical process could determine the form of a biological organ was an epiphany. She set about trying to find other examples and chose, as her model, avian skin. Chick embryos are easy to work with—they are a staple in developmental biology—and the follicles they develop closely resemble the ones from which human hair grows.
Previously, scientists hypothesized that the clumping behavior shown in Shyer’s stop-motion movie was driven by a unique gene expression pattern that caused cells to congregate around polka-dot concentrations of proteins. One particular protein, beta-catenin—which, among other things, helps cells adhere to one another— was thought to coordinate the entire process.
Recently, however, Shyer and co-author Alan Rodrigues, showed that avian skin cells need no such master regulator to begin rearranging themselves. When she removed beta-catenin from the picture, the cells still happily formed little clumps, so long as the membrane they rested on was, like the bed in the story of Goldilocks and the Three Bears, neither too hard nor too soft.
Further experiments revealed that, once this clustering of cells was under way, it caused beta-catenin to accumulate in the cells’ nuclei, presumably triggering the gene-expression changes required for follicle formation to proceed. These findings added considerably to our understanding of skin development, advancing a narrative in which beta-catenin, though still important, was no longer the all-powerful biochemical puppet master. Shyer and Rodrigues also illustrated how mechanical events can sometimes drive molecular ones, and not the other way around.
Shyer and Rodrigues, a senior staff scientist in the lab, suspect that in nature, embryonic follicle development probably involves a continuous give-and-take between mechanical and molecular processes, with changes on one side triggering responses on the other. Elucidating the precise sequence of mechanical and molecular steps in that feedback loop should help researchers understand human skin morphogenesis and enable them to grow more realistic skin tissue in the lab for research purposes.
In addition, the Shyer lab’s work may spur the development of powerful tools for studying and treating disease. Their discoveries, for example, could lead to improvements in organoids, small lab-grown simulacra of human organs such as brains and livers that hold great potential for biomedical research and regenerative medicine. These ersatz mini-organs might one day allow researchers to more effectively study the development of diseases and to test new drugs more realistically than can be done with rats and mice.
There could be other payoffs as well. Rodrigues and Shyer are currently investigating the mechanical underpinnings of tumor formation in hopes of developing novel strategies for treating cancer—strategies that look beyond the bewildering assortment of genetic errors that can cause cells to turn cancerous and instead address the mechanical processes by which tumors grow and evolve.
“Cancer is fundamentally a physical problem, and it’s related to how cells and tissues behave,” Shyer explains. As a result, drugs that target the molecular mistakes that generate cancer cells could be even more effective if they were combined with treatments that addressed the physical events involved in tumor development.
“Mechanics can’t just be a side dish to the way we think about biology and development,” she says. “Its power lies in how we can connect it to what we know about how genes are expressed and regulated.”
Shyer’s findings raise an important question: If mechanical forces can induce cells to change behavior, how do cells detect these forces in the first place? In other words, how does a cell “feel” when it’s being nudged by a neighbor, or whether the surface it’s resting on is squishy or stiff?
Gregory M. Alushin, head of the Laboratory of Structural Biophysics and Mechanobiology, is attempting to solve these mysteries with a combination of approaches, including a cutting-edge form of electron microscopy.
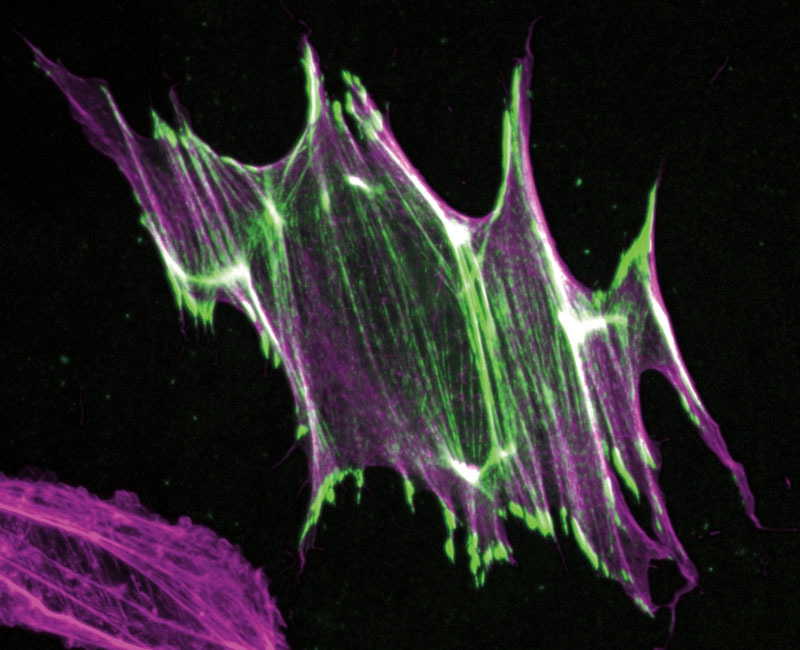
Alushin studies mechanosensation, the process in which cells use their internal cytoskeleton to detect the forces they experience.
When a cell finds itself on a rigid surface, or when it comes into contact with another cell, its cytoskeleton is deformed. This communicates important information to the cell (“Warning: stiff floor ahead”; “Ugh, it’s getting crowded”), triggering a cascade of biochemical signals and initiating an appropriate response.
In many cases, the response is movement. The cell’s motor proteins—specialized molecules that can produce mechanical force and move under their own power—tug on bits of the cytoskeleton, altering its shape, shifting its balance, and setting the cell in motion. Ultimately, the cell begins to migrate.
Cell migration happens during healthy tissue development as well as in some diseases, including cancer. The more rigid a breast cancer tumor becomes, for example, the more likely the cells composing it are to migrate—which, in turn, increases the likelihood that the cancer will spread to other organs and decreases the patient’s likelihood of survival. Understanding the mechanical processes involved, and the biochemical signals they elicit, could conceivably lead to new drugs for fighting cancer and spurring tissue regeneration.
But scientists don’t yet understand exactly how changes in the cytoskeleton allow the cell to sense that it is being subjected to mechanical force. Nor have they identified the various molecular actors involved in converting, or transducing, mechanical signals into biochemical ones. They also have yet to determine exactly how a cell goes from experiencing a tug or a push to ramping up the production of a protein or crawling in a particular direction.
“We’re focused on where molecules go and how they physically transform in response to mechanical forces,” Alushin says. “That transformation could be at the level of ‘turn left while you’re moving’ or at the level of ‘let’s change the expression of this or that gene.’ You need to understand those signals at the molecular level in order to design drugs that can interfere with them.”
Alushin is trying to achieve that by studying filaments made of the protein actin—thin threads that make up a major component of the cytoskeleton. He uses modified motor proteins to stretch these filaments to the point where they begin to rupture. He then takes high-resolution pictures of the filaments using cryo-electron microscopy—a technique that involves flash-freezing samples with liquid ethane, bombarding them with beams of electrons—and analyzing the resulting images using powerful software algorithms.
7 nmThe length of a typical actin filament.
In so doing, Alushin has identified lesions on the filaments that he believes attract specific molecules—including a protein, FHL, that binds to actin in response to force. That last discovery promises to be highly significant because FHL molecules also happen to regulate gene expression.
Previous research has shown that FHL shuttles into the nucleus when cells are exposed to soft environments, and it shuttles out when they are in stiff ones, toggling gene expression on and off in response to differences in force. As in the case of Shyer’s findings in avian skin cells, this appeared to be an abject lesson in how mechanics can influence biochemistry; but the mechanism driving that process remained unknown.
Alushin therefore engineered FHL, coupling it to fluorescent green protein so that he could track it using fluorescence microscopy. He then placed cells containing this modified FHL in a cell-stretching machine—a device resembling a wine fridge that stretches cells out like microscopic prisoners on a high-tech version of the medieval rack. (More precisely, the machine stretches the flexible silicone-based chambers in which the cells are deposited; but since the cells adhere to the surface of the chambers, they get stretched too.)
The resulting images clearly showed that FHL responded directly to force, coating a cell’s actin filaments or accumulating in its nuclei depending on whether it had been stretched. Alushin suspects that FHL binds specifically to the lesions he previously identified on purified actin filaments, and he plans to test that hypothesis by using cryo-EM to image the two in flagrante delicto both inside and outside cells.
With new compounds, it might become possible to stop diseased cells from moving to the wrong places.

“That would reveal the physical mechanism for this actin-recognition ability and could potentially help others design molecules to promote or interfere with it,” he says—establishing a “launching pad” for drugs of various kinds. With such compounds, it might become possible to encourage healthy cells to grow and move to the right places, for instance, or stop diseased cells from proliferating and moving to the wrong ones.
Alushin’s cryo-EM setup can’t yet provide quite the level of detail he requires. But he is already upgrading it—adopting new hardware for imaging entire slices of cells, for example, and developing new methods for boosting the resolution of his images—to push the quality of his data to the point where he can determine precisely how FHL binds actin, and how mechanosensation occurs in real life.
“I think you’re going to see more and more of this,” Hudspeth says of such technical innovations. “People will be asking mechanical questions and answering them with novel apparatus, because all of this is uncharted territory.”
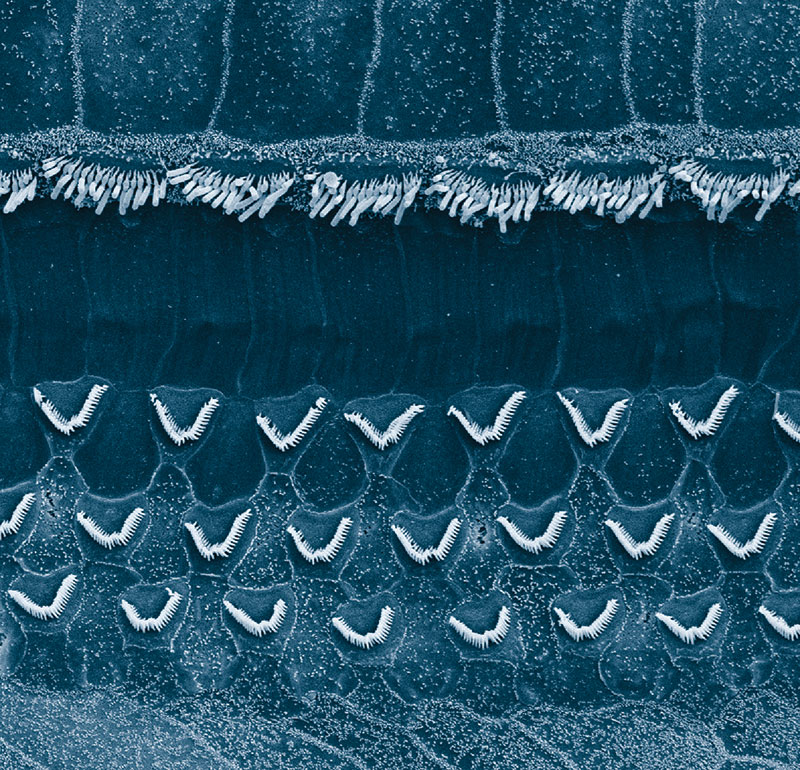
For more than four decades, Hudspeth, the F.M. Kirby Professor, has investigated the physiological basis of hearing—and he has the props to prove it. His office is littered with homemade devices for demonstrating how hair cells, the primary auditory receptors in our ears, transduce mechanical sound waves into electrical signals that can be interpreted by the brain.
“At a cellular level, this is the most complicated biological machine there is,” he says, grabbing one device after another to illustrate his point.
“You’re going to see more and more people asking mechanical questions and answering them with novel apparatus. All of this is uncharted territory.”

A metal sculpture in the corner depicts a hair bundle: a densely packed bunch of actin-filled fibers called stereocilia that extend from the surface of every hair cell. A hinged contraption represents a mechanically gated ion channel that opens and closes as the stereocilia sway back and forth in response to physical vibrations, generating electrical currents. And an exercise band stands in for the protein filaments called tip links that extend from the stereocilia to the ion channels, sliding them open as the stereocilia bend away from the channels and closing them as they do the opposite.
Hudspeth and his team believe that these tip links are a crucial component in the hearing system’s internal amplifier: a physical mechanism that boosts incoming auditory signals like a built-in hearing aid. When functioning properly, this amplifier allows us to hear the proverbial pin drop. But when it is damaged or degraded by injury, illness, or age, hearing loss ensues. Divining its secrets is therefore vital to helping the hundreds of millions of people who suffer from hearing problems.
For decades, scientists have suspected that a so-called gating spring endowed with elastic properties must be involved in opening and closing the ion channels located at the base of the stereocilia. Elasticity is key, since if the gating spring were too stiff—more like a pencil than a rubber band, for instance—our hearing system would lack its extraordinary sensitivity. Tip links have long been thought to serve as gating springs, but that theory has not been without controversy, in part because tip links are composed of relatively stiff proteins called cadherins that some argue are not suitable for the job.

Recently, however, Hudspeth and his team proved that this stiffness might not be a problem after all.
Doing so required another piece of cutting-edge technology: so-called optical tweezers, which use tightly focused laser light to trap and manipulate individual molecules. More specifically, it required an extraordinarily precise set of optical tweezers custom-built by postdoctoral associate Tobias Bartsch.
“There’s probably no more sensitive microscope on the planet,” Hudspeth says of the device, an unruly looking assemblage of cables, mirrors, and mysterious black boxes that lives in the basement of the Bronk building, isolated from stray vibrations that might throw off its exquisitely accurate readings.
85 dbThe point at which a sound may start causing permanent hearing damage. (Calm conversations are around 60 decibels.)
For their experiments, Hudspeth and Bartsch tethered each end of a string-like cadherin molecule to two tiny beads, one fixed, the other mobile. The researchers then used Bartsch’s optical tweezers to move the mobile bead by irradiating it with a laser, stretching the cadherin out with various amounts of force and measuring just how far and how fast it was able to extend. Those measurements, which were accurate to a single nanometer, or one billionth of a meter, proved the protein possesses mechanical properties compatible with the idea that tip links function as gating springs.
Now the team is trying to determine whether tip links are physically capable of transmitting mechanical force between stereocilia and ion channels fast enough for hearing—no mean feat given that humans can hear sounds at frequencies of up to 20,000 cycles per second, requiring response times as small as nanoseconds.
At the same time, the researchers are using their optical tweezers in conjunction with genomic methods to explore the mechanical consequences of the roughly 150 genetic mutations that affect tip link proteins. Though often extremely subtle, those mutations are nonetheless associated with a variety of developmental abnormalities, including deafness.
And in much the same way that Amy Shyer examines how the interplay between mechanical and biochemical events causes skin cells to form follicles, postdoctoral fellow Anna Erzberger, in Hudspeth’s lab, is investigating the combination of molecular and mechanical factors that drive the development of sensory organs in zebra fish.
“We all agree that biochemistry is only half of the story, and the other half is mechanics,” Erzberger says.
That sentiment is shared by many researchers. But perhaps nowhere is the complementarity between classical mechanics and 21st-century molecular biology more palpable than in the work of Shixin Liu, head of the Laboratory of Nanoscale Biophysics and Biochemistry. His work illustrates, among other things, how violent collisions between molecules play an important role in ensuring healthy gene expression.
Liu investigates molecular machines: specialized protein complexes that convert chemical energy into mechanical work. Measured in nanometers, these minuscule workhorses do much of the heavy lifting inside our bodies. The motor proteins that Alushin uses in his research, for example, belong to a class of molecular machines that contract our muscles, transport chemical cargo, and enable the skin cells that Shyer studies to crawl about.
“They’re not that different from real-world machines,” says Liu, who compares them to car engines, albeit ones that burn high-energy biological molecules rather than gasoline.
Liu is particularly interested in the molecular machines that power gene expression and regulation. These complex tasks require close coordination among several different molecular machines, including DNA and RNA polymerases, which replicate DNA and help manufacture RNA, respectively.
Scientists do not yet fully understand the mechanics, much less the consequences, of what happens when these molecular machines come into physical contact with one another, with our DNA, and with the chromosomes inside which that DNA is stored—in part because those interactions are so subtle that, until recently, scientists had no way of directly visualizing and recording them. Yet such information could prove invaluable.
“These machines are implicated in disease: cancer, neurodegenerative diseases, and many others,” Liu says. Consequently, understanding how they both enable and disrupt normal gene expression could lead to all manner of novel drugs and therapies.
To that end, Liu has built a unique experimental platform for analyzing the intricate ballet these machines perform together.
On the one hand, he uses high-resolution fluorescence microscopy to create movies of molecular machines as they bind to DNA and do their work. By tagging a machine such as DNA helicase with a fluorescent marker, for instance, he can watch as it alights on a piece of DNA, unwinds the double helix so that DNA polymerase can read it, and flits off again. By tagging different bits of the machine with different colors, he can deduce the conformation of the protein. By tagging two different machines with differently colored probes, he can see how they interact.
On the other hand, he employs optical tweezers like the ones Bartsch uses to poke and prod molecular machines in ways that reveal their underlying mechanics.
You can paint incredibly detailed pictures of how these machines operate on chromosomes—and see how their interactions can both harm and help us.
“You can pull them, you can twist them—you can see how these molecules respond to force,” Liu says.
That’s important, he explains, because the molecular machines and genetic materials inside our cells are constantly buffeted by forces of various kinds. Subjecting them to comparable treatment in the lab is therefore the only way to get a realistic idea of their mechanical properties.
By combining these two streams of data with next-generation genomic techniques, Liu can paint incredibly detailed pictures of how molecular machines operate on chromosomes—and how their mechanical interactions can both harm and help us.
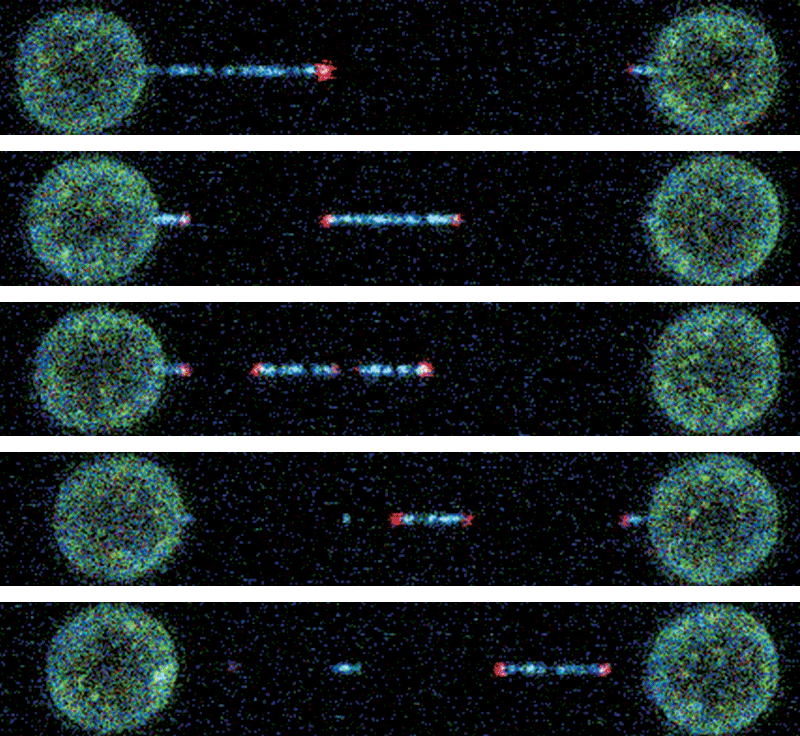
In the crowded environment of the cell, for example, DNA and RNA polymerase sometimes collide like runaway locomotives on the same track, causing genetic mutations that can lead to cancer.
In a recent study, however, Liu and his colleagues were able to demonstrate that at least some head-on collisions between RNA polymerases may actually serve a useful purpose; namely, preventing the molecular machines from reading too far into a genetic sequence.
“You want the gene to be stopped precisely at a well-defined position,” he says. Sometimes the stopping mechanism appears to involve a nanoscale pileup, and to flesh out precisely how this works Liu plans to observe such molecular train derailments in action.
Given the differences in scale and purpose, Liu’s molecular movies may seem but distant relatives of Shyer’s cellular slide shows, just as Alushin’s cell-stretching actin experiments may seem worlds away from Hudspeth’s optical tweezer tests.
Uniting all these projects, however, is a conviction that there is much to be learned from the physical interactions of biological components that cannot be gleaned by any other means—and that what has been discovered so far represents only the tip of the iceberg.
“The role of mechanics in biology,” says Hudspeth, “has been greatly underestimated.”