Feature
Life, illuminated
Innovations in imaging are unblurring our world, pixel by pixel.
By Eva Kiesler and Zachary VeilleuxScience, at its core, is observation. More than anything, we want to know what our universe looks like. That’s true for the world’s greatest experimentalists as well as for kids getting their first peek at a cell though a classroom microscope; we are visual creatures, and seeing is believing.
In the past 500 years, microscopes have evolved from crude pieces of glass capable of magnifying insects into multimillion-dollar instruments—precision-crafted machines that can illuminate, manipulate, record, and quantify the tiniest minutiae of life and disease. Even today, bioimaging continues to improve at breakneck pace, driven by advances in optics, biochemistry, electronics, and computing.
Here’s a sampling of images from the front lines of the field.
Super-resolution superpowers
At its most basic, microscopy is about using lenses to make tiny objects easier to see. For the first few hundred years, advances in the ability to shape and position glass drove most of the improvements in the field; in the 20th century, light microscopes acquired capabilities such as fluorescence imaging and optical sectioning, giving us closer and closer views of subcellular structures and macromolecules. But eventually, we came up against physics; there’s only so much detail you can capture before light waves are redirected away from the lens, rendering images hopelessly blurry. It’s a principle called the diffraction barrier, and because of it, most light microscopes max out at a resolution of about 200 nanometers (about one-fortieth the length of a red blood cell).
Eventually, we came up against physics: there’s only so much detail you can capture before light waves are redirected away from the lens, rendering images hopelessly blurry.
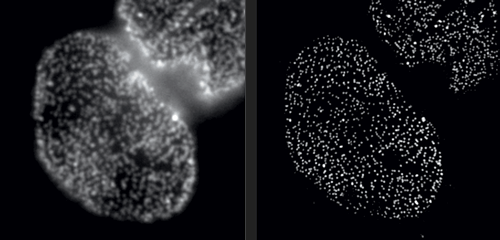
But in Rockefeller’s Bio-Imaging Resource Center, research associate professor Alison North demonstrates one of her latest acquisitions: a 3D super-resolution microscope, capable of circumventing the laws of physics. With this instrument, North and her colleagues are producing images that were impossible to obtain only a few years ago. Among them are crisp photos of nuclear pore complexes, protein assemblies that perforate the outer membranes of a cell’s nucleus. These structures, which are only about 120 nanometers in diameter, were previously doomed to fuzziness.
Super-resolution technology allows you to hack the diffraction barrier. The microscope superimposes a grid-shaped light pattern on a specimen and shifts the grid while capturing a series of images, which are then fed to a computer. “The patterned light interacts with the fine details to make them coarser,” North explains, “bringing the visual information into a range where we can collect it. And once we have the data, we can reconstruct the fine details using mathematical algorithms.”
Seeing by swelling
When even super-resolution doesn’t work, it’s time for plan B. Hironori Funabiki, who studies how cells segregate chromosomes during cell division, employs a surprisingly simple solution: he makes his sample bigger.
The result is an oversized cell that is anatomically correct, although its biomolecules are fragmented.
Pavan Choppakatla, a graduate student in Funabiki’s lab, uses a technique known as expansion microscopy to visualize the distribution of proteins that are thought to fold DNA into chromosomes. The method relies on a superabsorbent polymer, the stuff that retains liquid in diapers. When exposed to water, the material expands and can eventually reach up to one thousand times its original volume. Initially developed at MIT, expansion microscopy requires scientists to build the polymer within a cell, and anchor it to various attachment points before initiating a chemical reaction to trigger expansion.
The result is an oversized cell that is anatomically correct, although its biomolecules are fragmented. These fragments sustain the configurations the molecules had before expansion, but with everything spaced further apart. In effect, Choppakatla can quadruple the power of a microscope with this technique.
“Because it gives us an overall view of numerous molecules across an entire cell, this technique is ideal for studying how very long stretches of chromosomal DNA are individually organized into rod-like structures that can be moved apart during cell division,” Funabiki says. By labeling the proteins of interest, his lab gets the same images they might otherwise obtain but with four times the level of detail.
Cells in action
Here’s what you might try in order to find out if a neuron is doing something: Attach a specific fluorescent marker to it and illuminate it with a laser. When the neuron is active, it will brighten.
“Much of the data we collect from a sample is redundant. We don’t need it if what we are actually interested in is the activity of neurons.”
It works well when the neuron is sprawling on a glass slide. But what if you want to watch it in its natural milieu, deep inside the brain of a living organism? What if you want to study not just one neuron, but thousands? And what if you need to deduce precisely which of them are communicating, millisecond by millisecond?
Alipasha Vaziri, a physicist and neuroscientist, is building instruments and computational algorithms that can accomplish all these things, and more. Stationed in an all-black, windowless lab in the basement of Smith Hall, his group’s microscopes cut through enormous amounts of data in order to capture wide-ranging brain functions at single-cell resolution.
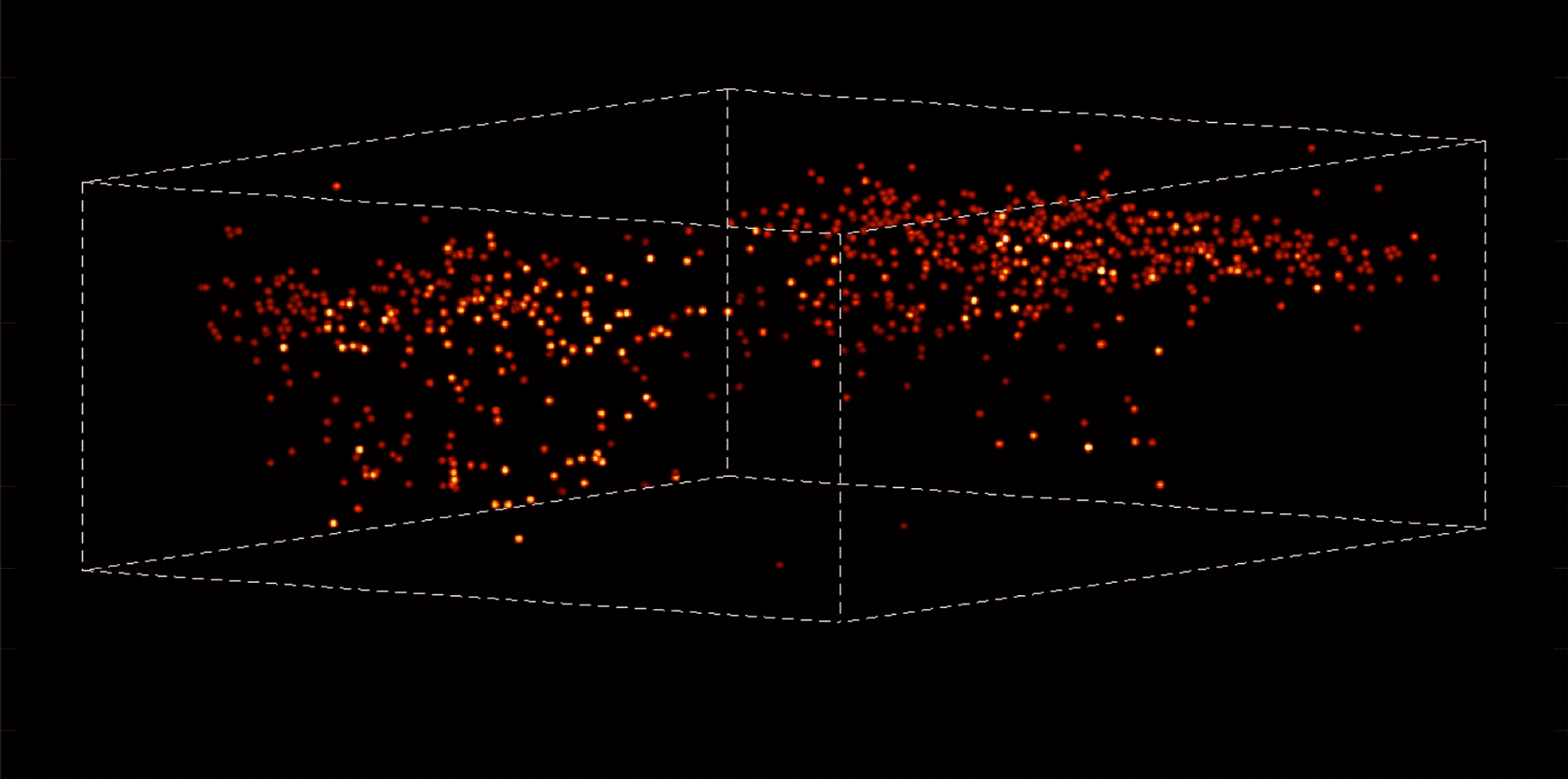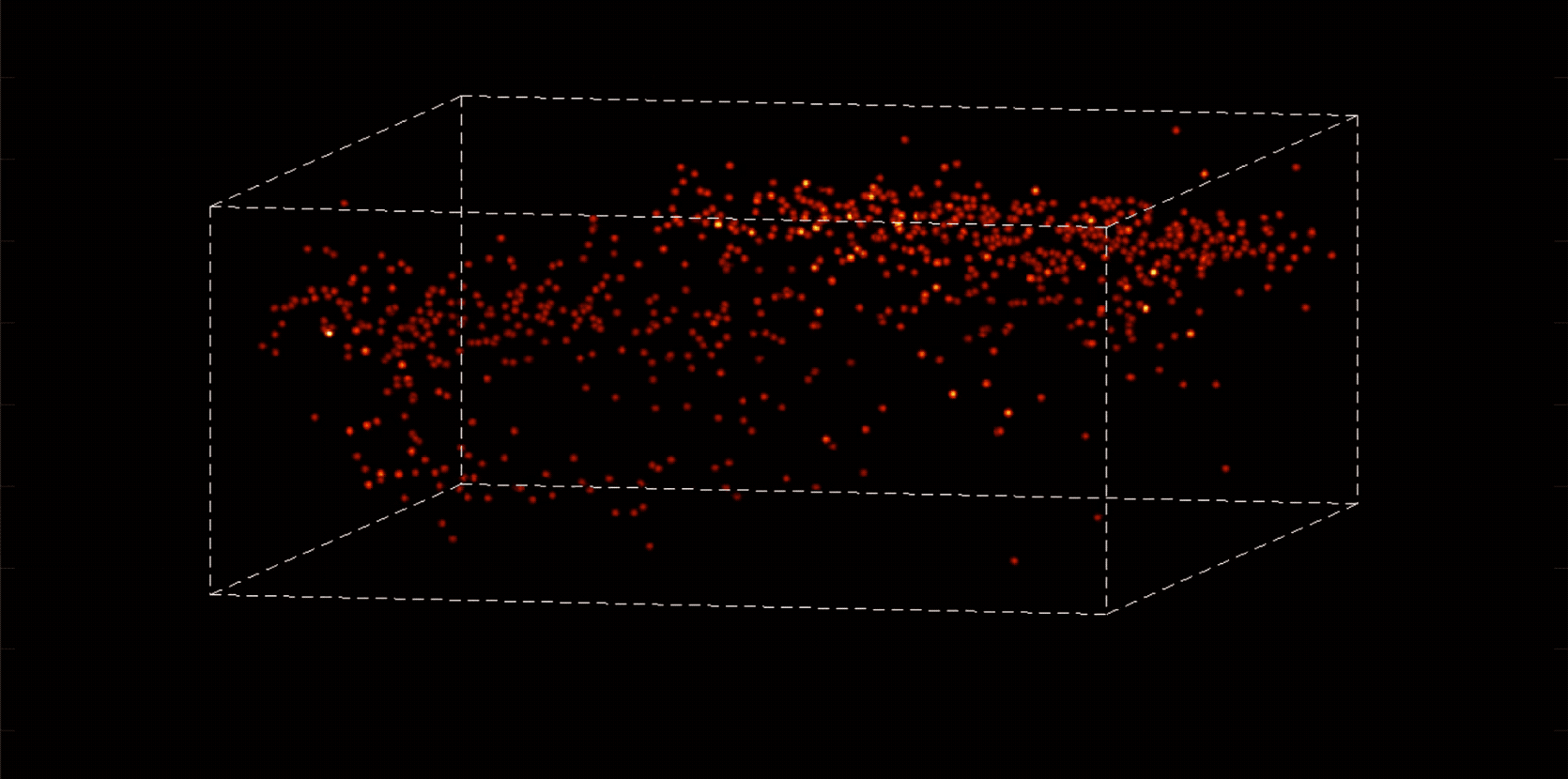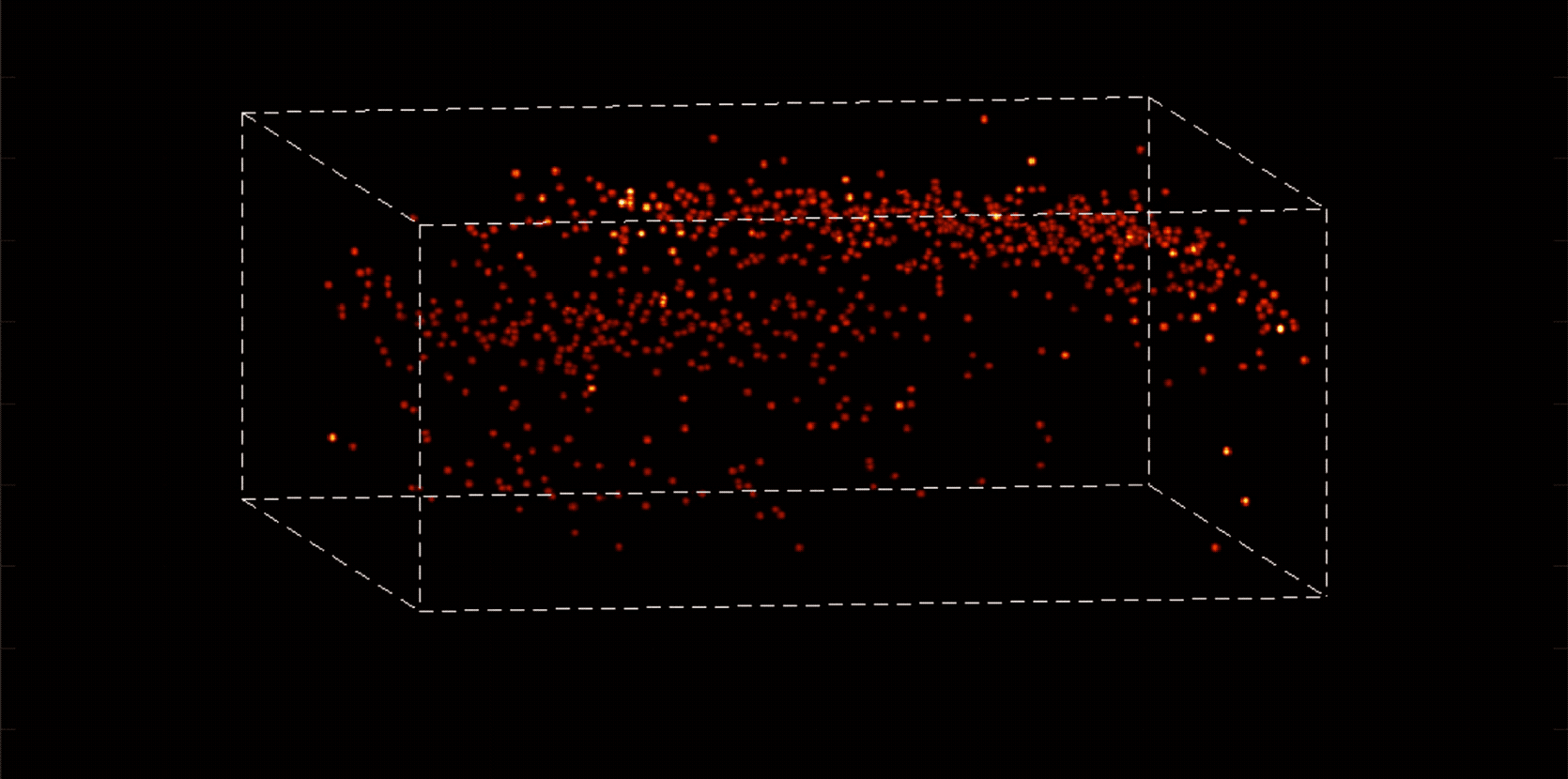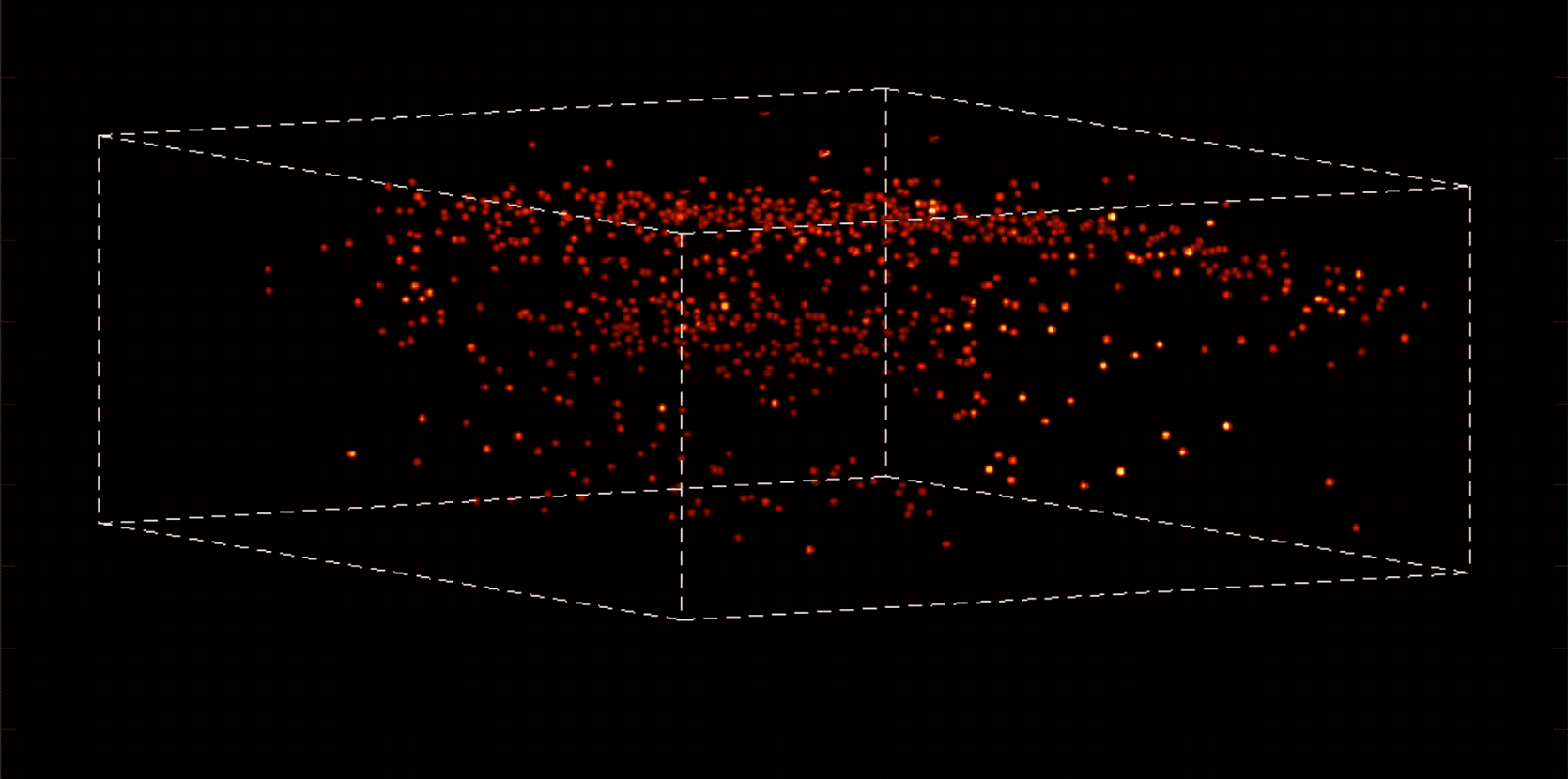
“With most traditional microscopes, much of the data we collect from a sample is redundant,” Vaziri says. “In neuroscience applications, for example, the size of the cells and their locations remain virtually constant over the duration of imaging—we don’t really need to acquire that information over and over again if what we are actually interested in is the activity of the neurons.” By putting the known information into a computational model, Vaziri is able to extract only the data that pertains to changes in the activity of nearby neurons from one moment to the next. The result is that less visible activities occurring within a mouse brain, for instance, become easier to see.
Essentially, Vaziri is using computation both to boost the sensitivity of the optics and to utilize that sensitivity to maximum effect. “We’re asking our algorithm: Given the patterns we are observing, and what we know about our sample and our equipment, what is the most likely position of neurons and their activity in time?”
The method can be used to compensate for the tendency of light to scatter as it travels through semi-opaque tissue, yielding striking moment-to-moment snapshots of neural activity. It can do it at high speed and over large areas of the brain, making it possible to capture an overall picture of activity patterns in even relatively large brains, like a mouse brain.
Worms on the move
There’s another challenge with imaging living things: They tend to be delicate. Even the most powerful imaging system will be of little use if it squashes or distorts the very thing you’re trying to look at. It’s a problem that Wolfgang Keil, a postdoc working in the labs of Shai Shaham and Eric D. Siggia, is acutely aware of.
Keil, who studies how organs and neural circuits develop in C. elegans, has developed a new way to get his squirmy subjects camera-ready. Traditionally, the flea-size worms are glued to a microscope slide when imaged, to keep them under the lens. It’s an unnatural setup that causes numerous problems: A trapped worm will soon get stressed, hungry, or hurt, so the microscopist must hurry.
To be able to image worms over long periods, Keil built a microfluidics chamber in which his C. elegans roam around and eat freely, except during short photo ops. When it’s time to take a picture, Keil gently draws a worm to the edge of the chamber, then lowers a ceiling to hold it still. Seconds later, the worm is free to go about its business—until the next snapshot.
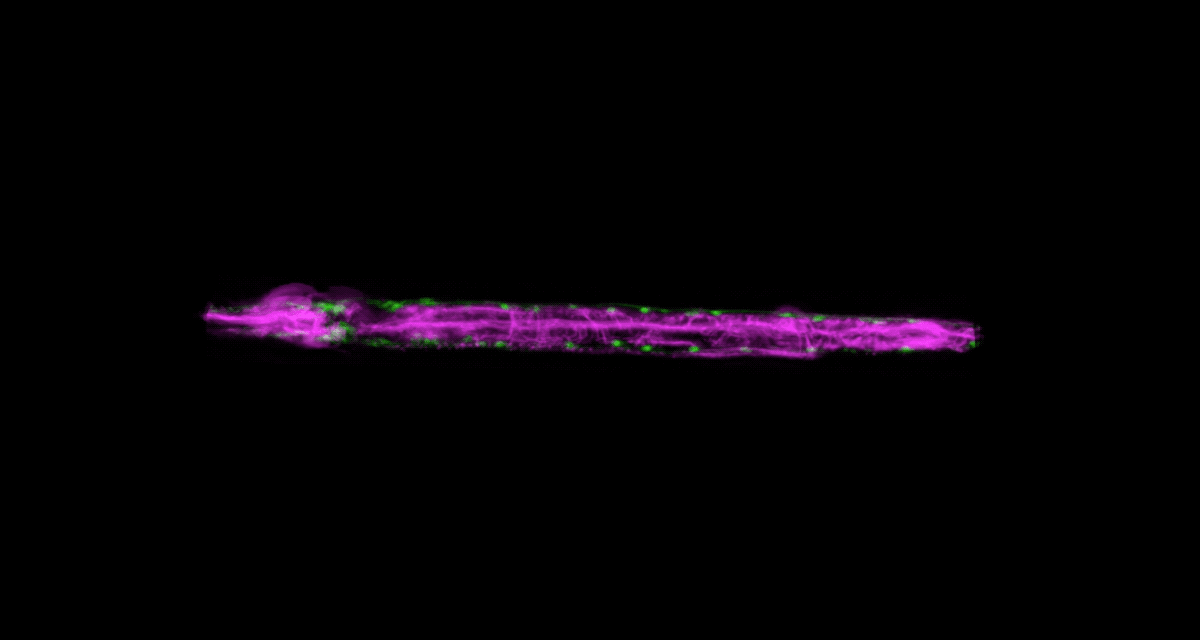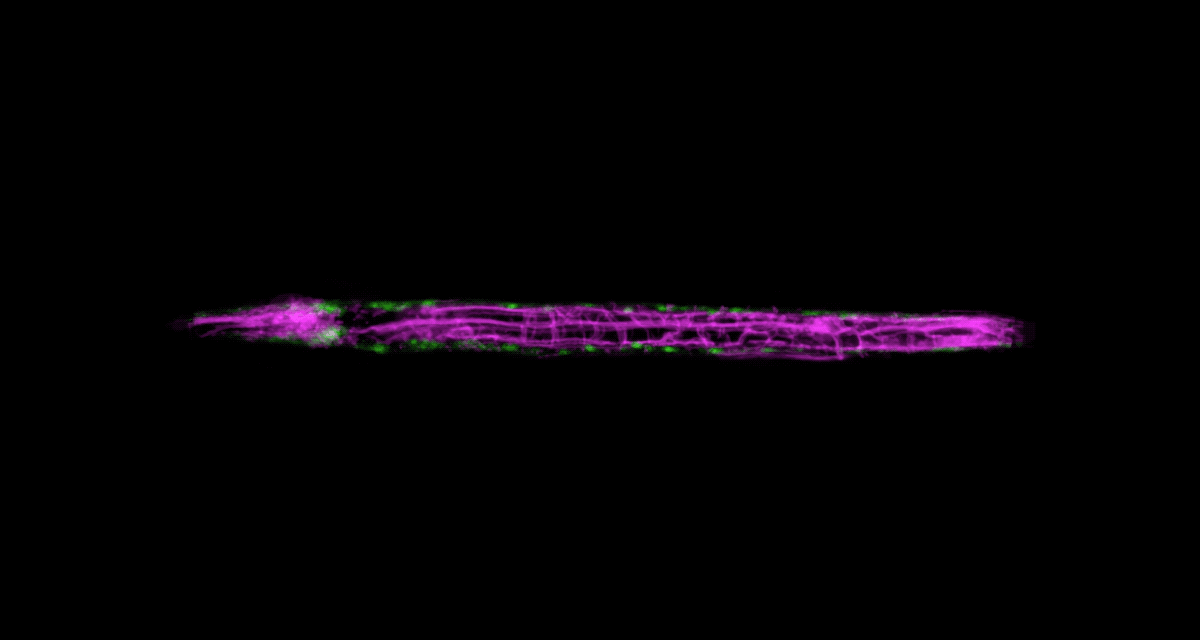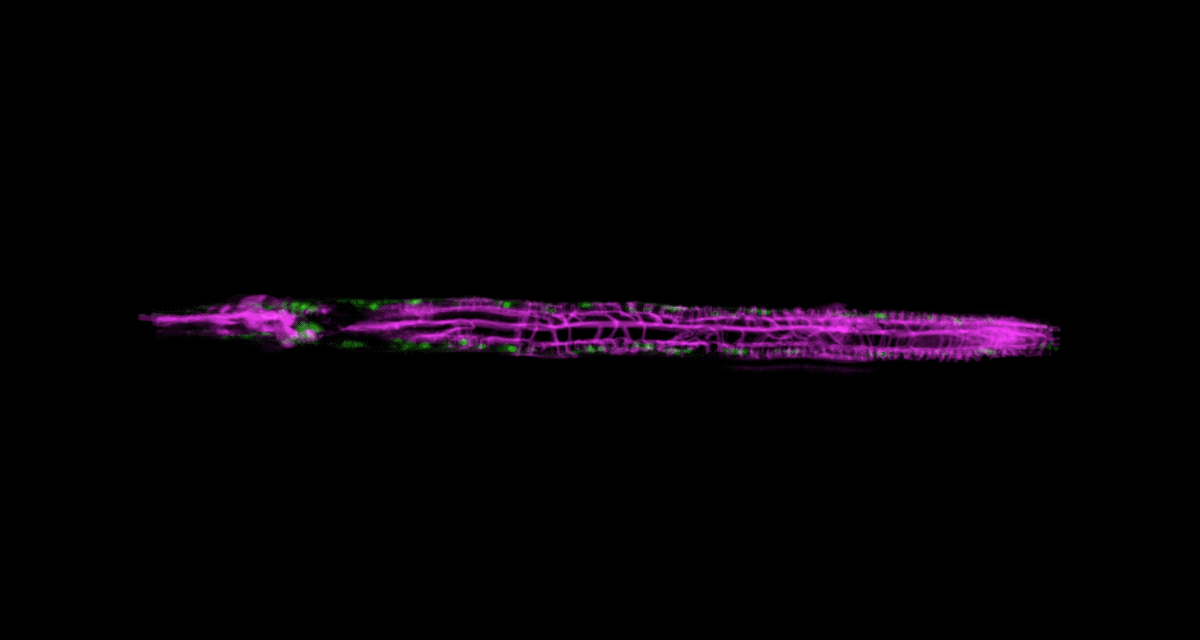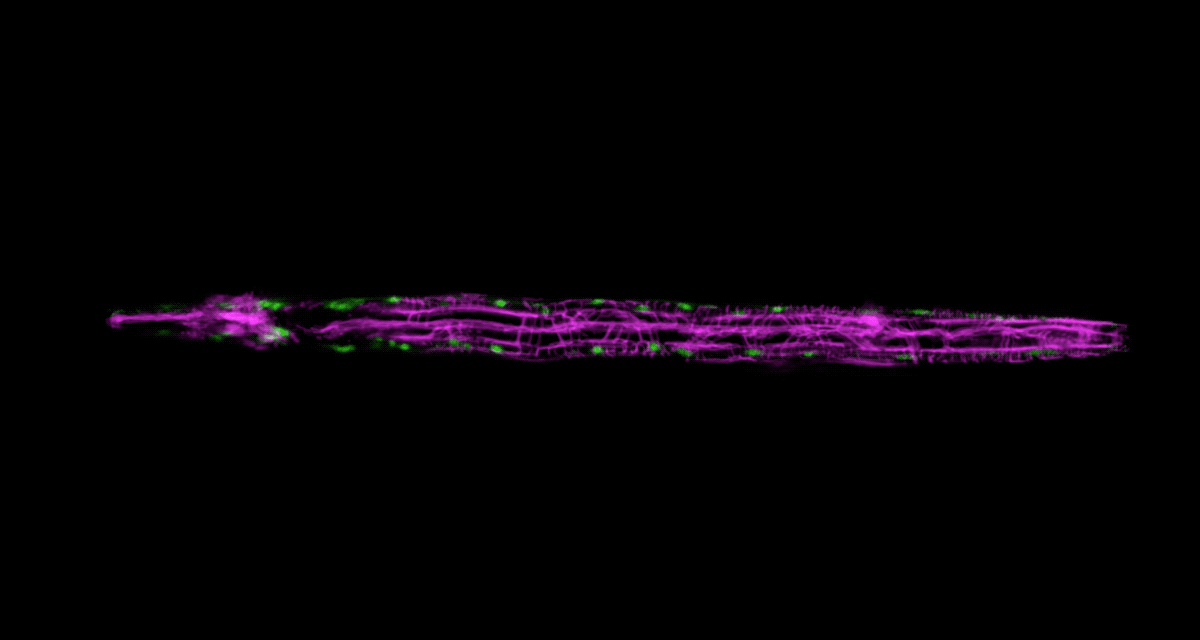
“The chamber has enabled us to do something that’s never been done before: to study a developing animal that’s feeding, growing, and interacting with its environment,” Keil says. Post-processing software can line up the worm snapshots in precisely the same orientation no matter which way the worm is facing, so the resulting time-lapse movies give a clear view of changes over time.
Because Keil’s system can image 10 worms at once, the method improves greatly on the statistical power and efficacy of previous ones. “The field has been completely lacking this capability,” says Shaham, the Richard E. Salomon Family Professor, “so people missed a lot of phenomenology.”
In a test drive, his team used the chamber to track four events in the development of C. elegans larvae. “In every single stage, we discovered new biology that hadn’t been described before,” Shaham says.
Cellular smoochers
If some biologists seek to prolong their imaging sessions, others are more concerned with speed. Gabriel D. Victora, the Laurie and Peter Grauer Assistant Professor, studies fleeting interactions between immune cells—encounters so brief that he refers to them as “kiss-and-run.”
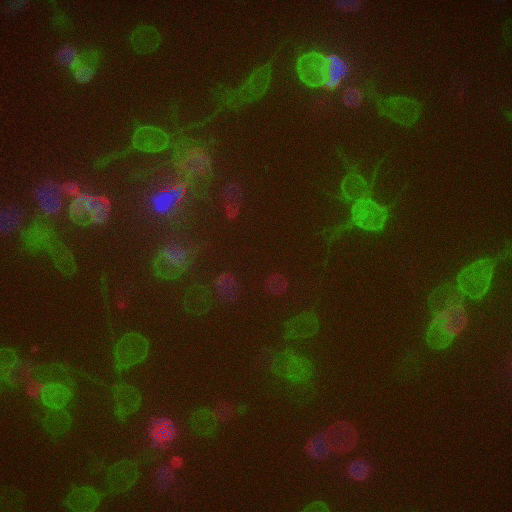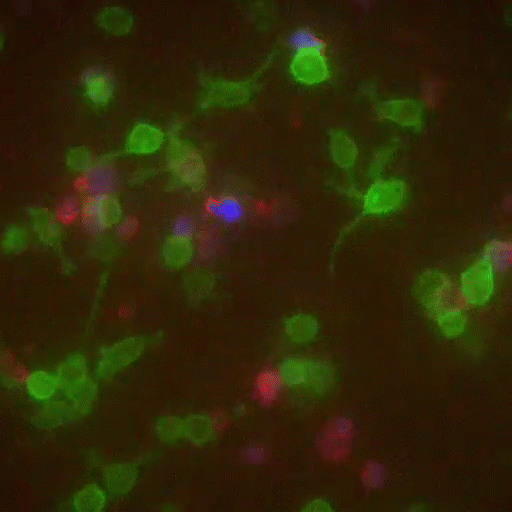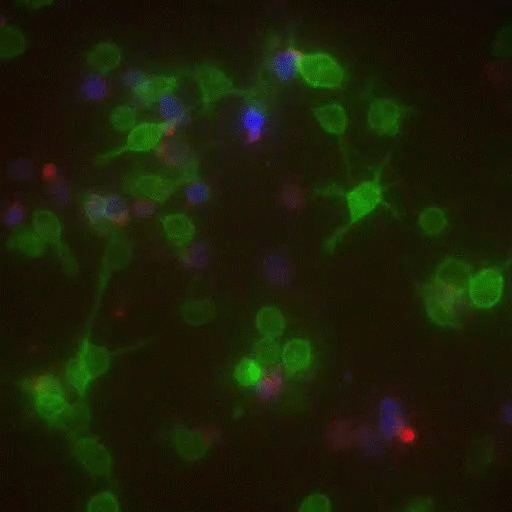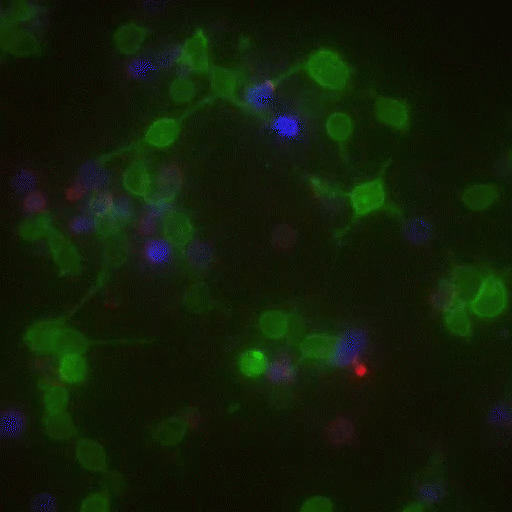
In a crowd of cells that look more or less the same, an occasional one will suddenly run up to a peer and make contact, then bashfully move away. “Virtually all of immunology is based on these exchanges,” says Victora, “in which two cells exchange signals to kick-start a response against a pathogen.”
Scientists previously examined these events only inside Petri dishes, but Victora has devised a way to capture them in living mice. It involves injecting the animals with immune cells engineered to produce a fluorescent marker—the biological equivalent of lipstick—then tracking the marker as it travels. Every time an immune cell kisses another, it smears it.
The spectacle is fascinating to watch under the microscope, and it’s also well-suited to detection via flow cytometry, which allows Victora to quickly count interactions within a big cell population. “This is how we figure out precisely which immune cells interact in a given scenario,” he says, “and how their communication changes over time.”
