Feature
A great chill has unlocked biology
Thanks to ice, structural biology is on fire. With new ways to snap-freeze and visualize molecules mid-movement, scientists are churning out new knowledge about life and disease with unprecedented speed.
By Alexander GelfandIt looks like a fancy espresso maker,” says Mark Ebrahim, gesturing towards a finely machined assembly of metal components housed in the basement of the Collaborative Research Center.
But Ebrahim is no barista. Rather, he is a physicist. And as the safety sticker on it suggests (“Warning: Cold Surface”), the device before him is no coffee maker.
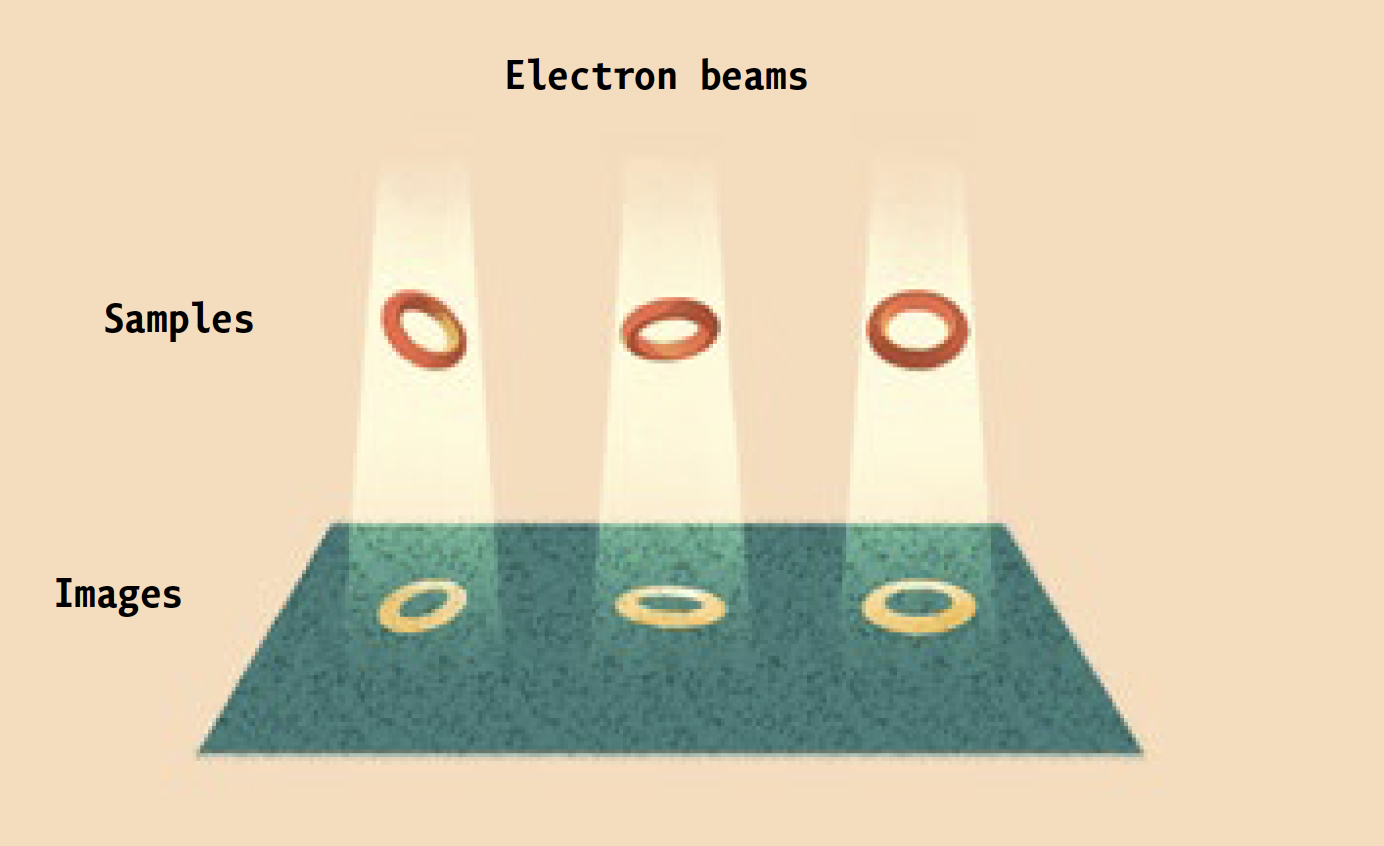
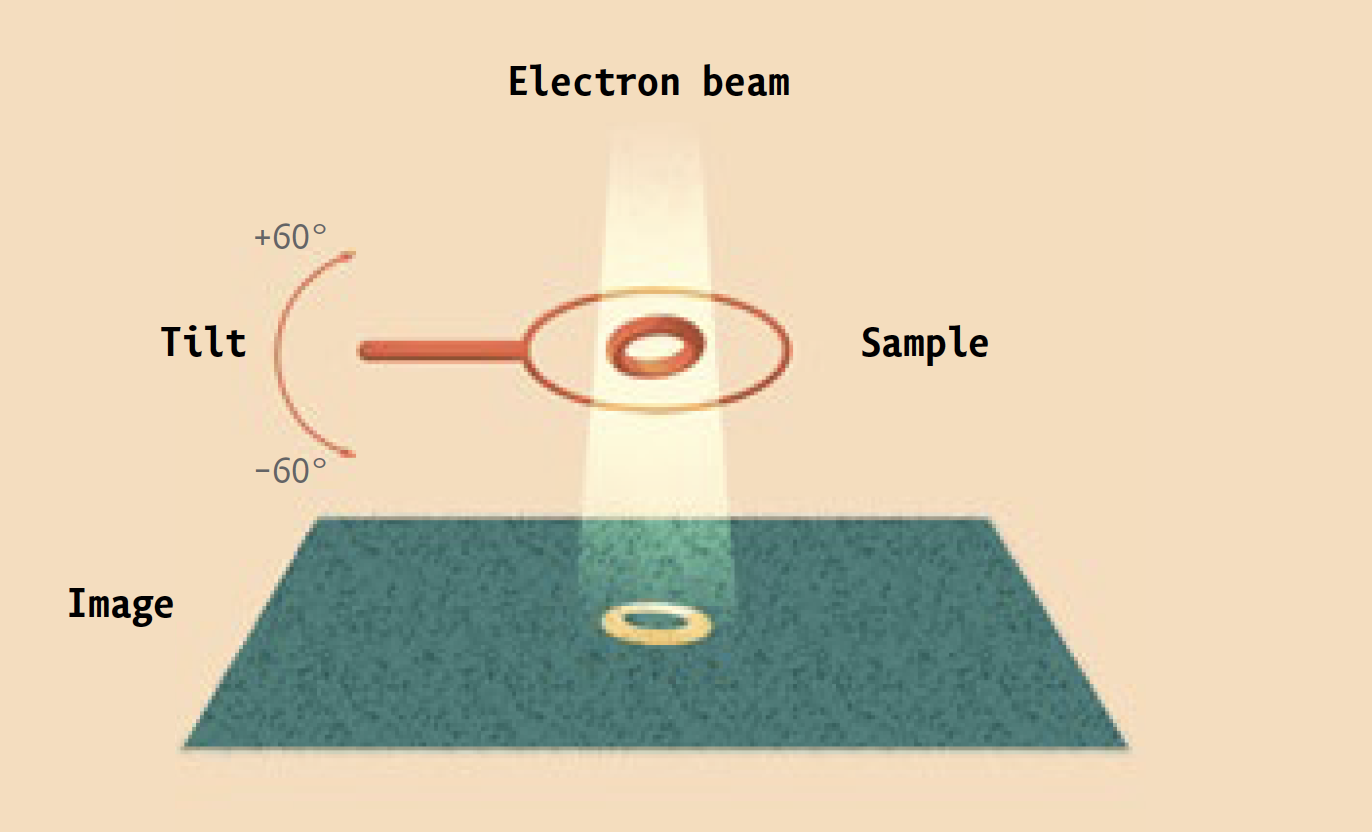
Instead, it is a high-powered electron microscope that pumps focused beams of charged particles through protein samples that have been flash-frozen in liquid ethane, and then kept at a chilly -180°C by the contents of huge liquid-nitrogen tanks. Super-sensitive cameras record the electrons that pass through the samples, and powerful software algorithms use the resulting data to generate three-dimensional images of the proteins at near-atomic resolution. (Ebrahim, a senior staff scientist, keeps the equipment running.)

The technique is known as cryo-electron microscopy, or cryo-EM. And it is enabling researchers in labs across campus to investigate biological molecules, and answer scientific questions, that had previously been off limits.
“A lot of impossible projects have become feasible,” says Jue Chen, the William E. Ford Professor. She speaks from experience: her own recent cryo-EM investigations are galvanizing research into cystic fibrosis, a debilitating lung disease for which there is as yet no cure.
When it comes to understanding the basic mechanics of life, structure is everything: the shape of a biological molecule dictates what the molecule does and how it does it. Until recently, the gold standard for determining the physical structure of proteins—the molecular workhorses of the body, responsible for performing a nearly endless variety of chores at the cellular level—was x-ray crystallography, a technique that involves turning proteins into crystals and probing them with electromagnetic radiation.
It’s a powerful method, capable of resolving structures at the atomic level. But it is not without its drawbacks.
For one thing, most proteins move through different shapes, or conformations, as they go about their work. But only one conformation can be crystallized at a time, preventing researchers from analyzing the full range of shapes that allow proteins to do the jobs that nature has assigned them.

Worse, not all proteins are amenable to crystallization: some are too fragile to withstand the process, while others can’t be packed into orderly three-dimensional crystals. Yet x-ray crystallography is an all-or-nothing proposition: “You get great crystals, you get a structure. You don’t get crystals, you get nothing,” says Thomas Walz, who heads the Laboratory of Molecular Electron Microscopy.
Consequently, researchers were left with a vast collection of proteins whose mysteries seemed destined to remain impenetrable, including the so-called membrane proteins that, among other things, help ferry vital substances in and out of cells.
Cryo-EM, however, changed all that.
Prior to being frozen, proteins destined for the electron microscope are comfortably suspended in a liquid solution that resembles their native environment inside cells, helping to preserve their integrity. Since many different conformations of the same protein can be trapped in ice simultaneously, scientists can see all of them at once.
“It gives me a view of the dynamics of the protein or protein complex I’m interested in,” explains Walz—who, when he isn’t lending his expertise in cryo-EM to colleagues in other labs, employs the method himself to explore the interactions between membrane proteins, other biological molecules, and the cell membranes they inhabit.
Despite these advantages, cryo-EM was not an overnight success. Originally developed in the 1980s, the technique initially produced such fuzzy, low-res images that it was jokingly referred to as “blobology.” But in recent years, improvements to the electron detectors and image processing software that make cryo-EM possible have vastly improved the resolution it can achieve.

As a result, the technology has enjoyed soaring popularity and newfound acclaim—three of the scientists who developed cryo-EM for biology were awarded the 2017 Nobel Prize in Chemistry. And other researchers who once relied primarily on x-ray crystallography—including Roderick MacKinnon, head of the Laboratory of Molecular Neurobiology and Biophysics, who used it in the work that won him his own Nobel Prize, in 2003—have now shifted almost entirely to cryo-EM in hopes of cracking previously insoluble problems.
One such problem involved a membrane protein called cystic fibrosis transmembrane conductance regulator, or CFTR, whose structure Chen spent nearly a decade trying to resolve using x-ray crystallography. After failure upon failure, she finally succeeded just last year, thanks to cryo-EM.

Number of Americans infected with drug-resistant Staph superbugs each year.
CFTR allows chloride ions to travel across cell membranes, and researchers have long known that mutations in the gene that produces the protein lead to cystic fibrosis, a chronic disease. (Defects in CFTR cause mucus to accumulate in the lungs, with potentially fatal consequences.) They did not, however, know what kinds of changes those mutations caused in the physical structure of CFTR, or how those changes prevented the protein from operating properly—information that could potentially unlock new treatment strategies.
Chen, who heads the Laboratory of Membrane Biology and Biophysics, began working on CFTR in 2008, but try as she might, she could not persuade the protein to crystallize—in part because a section of it, known as the R domain, doesn’t have a fixed structure. It flops about from one position to another.
A few years ago, Chen began learning the ins and outs of cryo-EM with help from old hands like Walz. She initially used the technique to study a form of CFTR found in zebrafish that is virtually identical to the human version; it was easier to work with, and allowed her to refine her methods. Thanks to those preliminary studies, she was ultimately able to get a good look at the human version of CFTR, even determining the location (or locations) of its elusive R domain. Her results, which were published in the journal Cell this past March, help explain how a particular mutation interferes with the protein’s ability to function, information that could open the door to new therapies.
Most treatments today deal with the symptoms of the disease (mucus buildup, lung infections) rather than its underlying causes. The only FDA-approved drug that addresses a problem with CFTR itself helps less than four percent of cystic fibrosis suffers—those with a specific genetic mutation. The rest are currently out of luck.
Chen says that further cryo-EM studies could reveal more about the structure of CFTR, and how different mutations cause the protein to fail in different ways. This, in turn, would make it possible to design drugs that bring relief to far larger numbers of patients, something that would have been inconceivable not long ago.
Gregory M. Alushin, who joined Rockefeller last year as head of the Laboratory of Structural Biophysics and Mechanobiology, is also using cryo-EM to pursue projects that were formerly out of bounds.

Number of Americans who suffer from chronic wounds, often as a complication of diabetes or other disease.
Alushin investigates how cells sense mechanical forces: the constant pushing and pulling generated by their own movements and by the environment that surrounds them. These forces can lead a cell to change its motion—encouraging a healthy one to migrate from one position to another, for example, or provoking a cancerous cell to metastasize—and may even cause it to turn genes on or off.
Research has shown that long strings of protein called actin filaments play a crucial role in all of this. The proteins in the filaments, which are strung together in repeating patterns that resemble double-stranded spiral staircases, change shape in response to the forces they experience. Those changes in conformation invite other molecules known as signaling proteins to bind with them, triggering a series of biochemical events that ultimately lead to changes in a cell’s behavior.
Understanding precisely how the proteins in the filaments change shape, and which signaling proteins bind to them, could allow researchers to design drugs that would stop the process in its tracks, preventing tumors from metastasizing, or warding off unwanted changes in gene expression.
To achieve that level of understanding, however, Alushin first needs to determine the atomic-scale structure of the proteins. Until cryo-EM, that didn’t seem possible.
“You can’t use crystallography on them, because they don’t pack into crystals,” he says. And even if they did, he adds, the very act of locking the filament proteins into well-ordered crystals would make it impossible to observe the range of conformations that allow them to do their work.
Freezing the samples solved these problems, and by using specialized “motor proteins” to pull on opposite ends of the filaments as they freeze, Alushin’s team can even see how they deform in response to mechanical force.
Much to his surprise, Alushin found that rather than straightening out when stretched, the twisting, double-stranded protein structures comprising the filaments instead developed what looked like “squiggly” regions. He suspects that these deformations are caused by one strand breaking and wrapping around the other, and that the resulting change in shape serves as a target for signaling proteins. He is already conducting further cryo-EM trials to test this hypothesis.
“We know these signals are happening, but we don’t know which molecules are involved, or how they interact,” he says. “So we’re putting together the molecular cast of characters, and observing the conformational acrobatics they undergo.”
Of course, like any scientific technique, cryo-EM has its own limitations and challenges.
“We’re putting together the molecular cast of characters and observing the acrobatics they undergo.”
It doesn’t work particularly well on the smallest molecules. The image processing software it relies upon cannot always make sense of all the different conformations in a sample. And it generates so much data—multiple terabytes per project, Ebrahim estimates—that simply storing it all poses a serious challenge. Rockefeller has recently upgraded its high-performance computing capabilities to handle the needs of cryo-EM, and is beefing up its data storage capacity, as well (see “Processing power”).
Meanwhile, the technology continues to improve, promising even more impressive results to come. And Walz, who has been involved with cryo-EM almost from the start, continues to be amazed by the progress that has already been made since the bad old days of blobology.
“Every day I’m astonished by what can be done,” he says, “and by how easy it has become.”