Feature
Terrestrial treasures
Topsoil is teeming with secret bacterial recipes, some of which could become lifesaving drugs. Chemists are using new tools to dig deep into this hotbed of medical potential.
By Wynne ParrySean brady’s stainless-steel incubator contains no sand, no moldering leaves, no silently squirming worms. But open its glass door and inhale: there’s that unmistakable, warm scent of moist earth.
Stacked in neat towers on the shelves inside are plates fuzzy with colonies from which the earthy perfume emanates. These are not just any bacteria; they are—potentially—the key to an entirely new generation of medicines. Master chemists, they serve as manufacturing plants for an astonishing range of chemical substances normally produced by soil microbes. For Brady, a chemist himself, these dirt-derived compounds are full of potential, capable of yielding killer antibiotics, powerful cancer chemotherapies, or blockbuster anti-depressants.
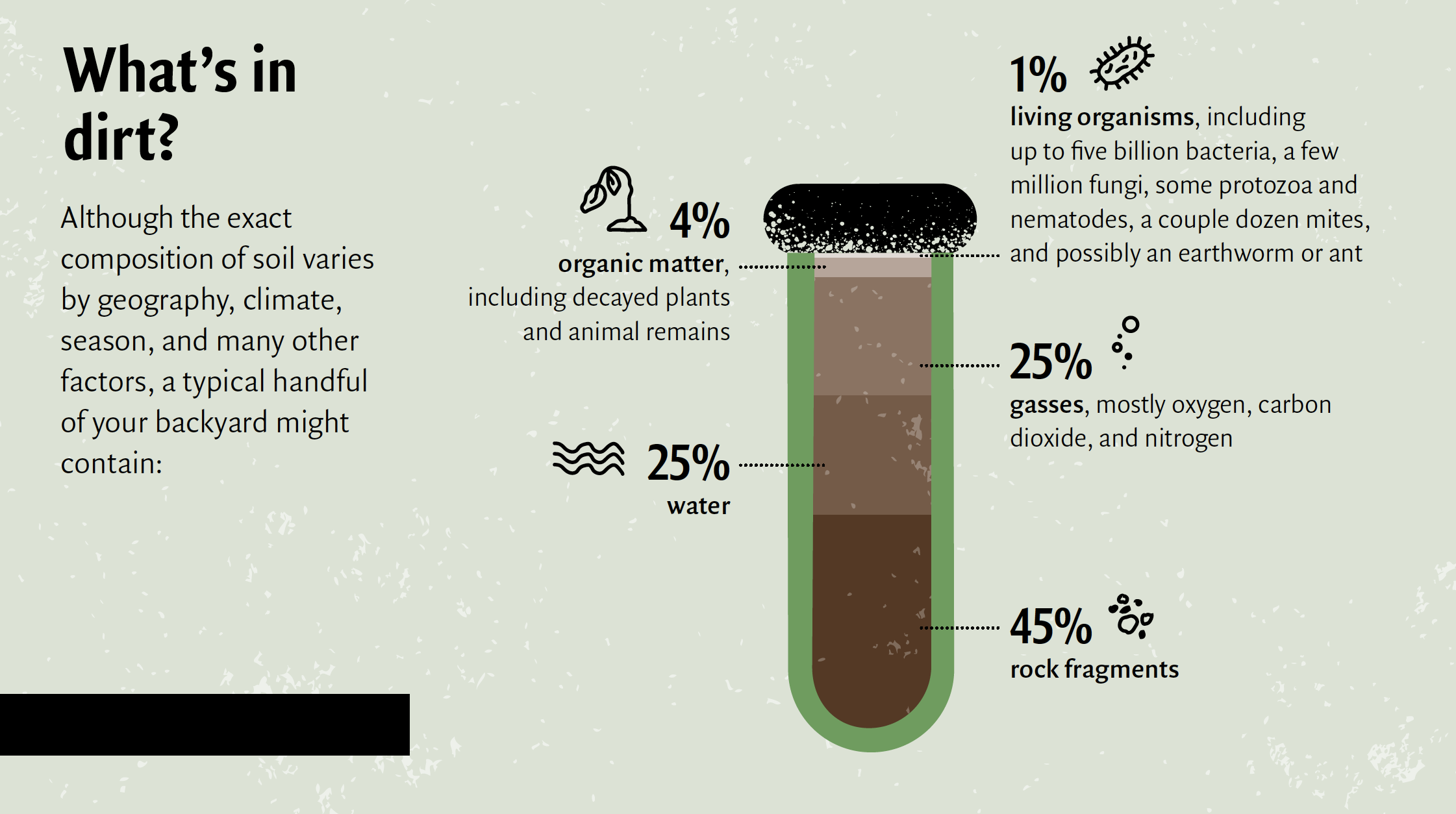
Soil is a logical place to look for obscure molecules: a single gram, less than a teaspoon, may contain thousands of species of soil bacteria—an immense source of untapped medicinal chemistry. “It’s the largest, most biodiverse reservoir of bacteria on the planet,” says Brady, “and it’s very easy to get.”
Dirt microbes are not easy to work with in the lab, however, and much of their pharmaceutical power has long been beyond human reach. But Brady, whose team has spent ten years developing new methods to mine soil for new molecules, is at the forefront of a drug renaissance.
His approach, a calculated combination of microbiology, chemistry, and computational biology, has opened the door to a world of chemistry no scientist has seen before.
Brady is not alone in his quest for esoteric soil substances. In fact, the ground beneath our feet is an ancient pharmacy that humans have exploited since long before we peered through our first microscope. Archaeologists, for instance, have found traces of tetracycline, an antibiotic produced by soil bacteria, in 1,500-year-old human bones from Sudan, suggesting that our North African ancestors were brewing and ingesting the drug on a regular basis.
The modern effort to produce this and similar compounds kicked off after 1928, when the doctor and scientist Alexander Fleming identified penicillin from airborne fungi. Following his discovery, other scientists began extracting lifesaving medicines from microorganisms, mainly soil bacteria. Among them were actinomycin D, a chemotherapy ingredient deployed against several types of cancer; streptomycin, an antibiotic that became the first treatment for tuberculosis; and other antibacterial drugs used to treat infections from acne to malaria.
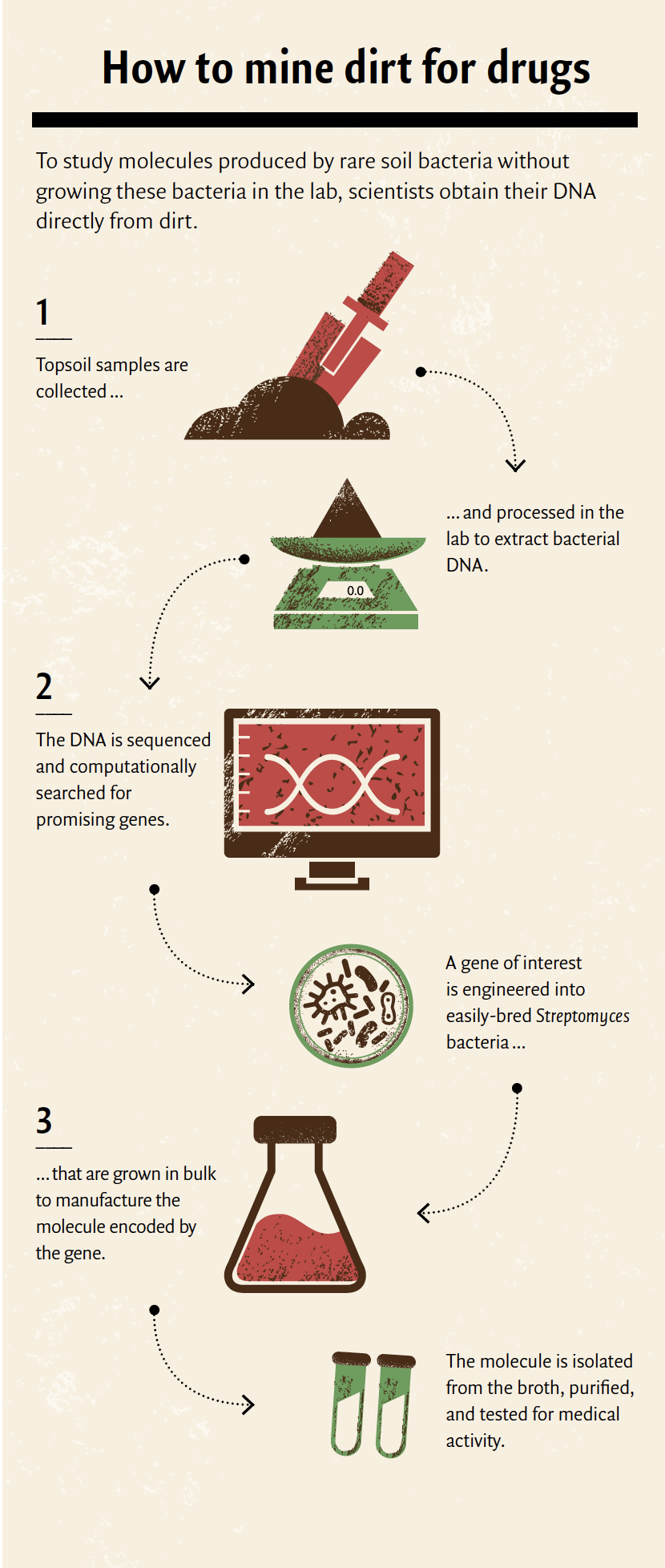
It’s now possible to extract a bacterium’s recipes for making chemicals without the messy, failure-prone process of actually keeping that bacterium alive.
Mid-century scientists were at the epicenter of an innovation boom that lasted a couple of decades—until the field ran up against an insurmountable obstacle. To be able to peruse a bacterium’s molecular contents, scientists first had to grow it in the lab, on plates or in shaker flasks; but most soil microbes won’t adapt to life inside an incubator. “The problem with growing unusual bacteria is that we don’t know what most of them eat, if they need companions of another species, or more space,” Brady explains.
So, from Fleming’s time until relatively recently, microbiologists had to concentrate their efforts on what turned out to be a relatively small number of domesticated species. To date, only a fraction of the world’s soil bacteria have been cultured—the rest are a black box.
Brady, who is the Evnin Associate Professor, has found a way to peer inside that box, however. Rather than trying to cultivate obscure and delicate microbes, his lab focuses on the soil itself, searching it for microbial DNA that contains the instructions for making new molecules. Thanks to new technology, it’s now possible to extract a bacterium’s recipes for making chemicals without the messy, failure-prone process of actually keeping that bacterium alive.

Among Brady’s peers, many have been successful in trawling bacterial genomes for drug-like substances using cutting-edge genome sequencing approaches. But Brady has set himself apart by looking not at the bacteria themselves, but rather at the DNA of organisms never grown in a lab.
“All Sean needs is DNA, and that’s the beauty of his approach,” says Jon Clardy, a professor at Harvard Medical School and the mentor who inspired Brady’s interest in the chemical-engineering potency of bacteria. “The strategy is conceptually very simple, but extraordinarily sophisticated from a technological standpoint.”
Think of it this way: if forensic scientists struggle to decode human DNA fragments hidden within, say, a piece of torn clothing from a crime scene, microbiologists like Brady face an even greater challenge examining DNA from dirt, which contains genetic material from thousands of unknown bacterial species—not to mention other subterranean beings like fungi, single-celled protozoa, microscopic worms, scraps of plants, and the occasional insect. To make progress, Brady has had to create new methods to find the DNA of interest, as well as brand new computational technologies to analyze it. Among other things, his lab uses highly specialized software to scan DNA sequence data derived from thousands of different soil environments for genes likely to produce agents with desirable qualities—molecules that mimic certain other molecules, for instance.
It’s a highly sophisticated approach that requires a litany of expensive equipment, technical know-how, and, to get things started, a shovel.

“Sample collection is easy,” says Zachary Charlop-Powers, a computational scientist and former postdoc in Brady’s lab. “The vast majority of microbes are found in the top 12 inches of soil, so we literally just dig.” He and his coworkers have collected dirt from the Rockefeller campus, from parks throughout the city, and from distant sites where traveling lab members have found themselves.
The work gets more complicated once the dirt arrives in the lab. Charlop-Powers and his coworkers extract bacterial DNA from it, and from there, the search may go in one of two directions. The scientists may conduct a targeted search for pharmaceutical molecules, guided by familiar chemistry (in one project, for example, they identified an agent similar to anti-cancer medicines called anthracyclines that they hope will be potent against tumors that no longer respond to those drugs). Or, in some cases, they take the “surprise me” approach, randomly testing all the molecules a bacterium can make for pharmaceutical activity.
Once they have identified promising pieces of bacterial DNA, the scientists move on to manufacture the corresponding molecules, a job that is in fact outsourced to a cooperative species of soil bacteria like Streptomyces, microbiology’s answer to the lab rat.
Reaching into the Brady lab’s fragrant incubator, Ian Woodworth, a research assistant, grabs a plate covered with orange-gold colonies of Streptomyces. Mixed in with their normal DNA, these bacteria harbor genes from other microbial species—molecular recipes that Brady’s team has found in a search for new drugs against tuberculosis. In particular, the researchers hope that some of these genes, now activated by the Streptomyces, will yield molecules capable of killing potentially deadly strains of Mycobacterium tuberculosis, the germ that causes lung infections.
Woodworth carefully picks Streptomyces colonies off the plate and releases them into broth-filled flasks, where the lab-friendly germs will be fed, aerated, and shaken for days. These colonies are essentially small chemical plants, pumping out their assigned molecules. (Of their own volition, Streptomyces also make geosmin, an oily molecule that gives these bacteria their earthy smell). From the enriched broth, Woodworth and his colleagues will later produce concentrated extracts of the compounds, which they will feed to tuberculosis bacteria. With any luck, the bacteria will die.
The same approach has led Brady and his coworkers to a number of molecules that they’re now developing as experimental drugs and testing in animals. Among them is what may be an entirely new class of antibiotics, one that appears frequently in soil samples collected across the country. These drugs, the scientists have found, have the potential to kill drug-resistant strains of Staphylococcus aureus, a germ that often causes dangerous infections in hospital patients.
“Sean has really been at the forefront of pulling relevant sequences out of the mishmash of DNA within soil,” says Jason Crawford, an assistant professor at Yale University whose lab, like Brady’s, uses genomics-based methods to discover new microbial compounds. “This has allowed him to do some wonderful things identifying and activating these natural products.”
In principle, brady’s approach could tip him off about new ways to treat nearly any type of disease. But there are good reasons why his lab pays particular attention to antibiotic drug candidates—molecules that look like they might have what it takes to kill germs in ingenious ways.
Despite having well over 100 antibiotics at their disposal, infectious disease doctors need more. Bacteria multiply quickly and are always mutating, which means the forces of evolution are keeping modern medicine on its toes: each time a new antibiotic is introduced, the microbes it targets begin devising ways to thwart it.
Health experts warn that humans are in danger of falling behind, and we may soon live in a world where antibiotics are of little use, and people die from common infections or minor injuries. Unsettlingly, a 2014 report by the World Health Organization states that “a post-antibiotic era … far from being an apocalyptic fantasy, is instead a very real possibility for the 21st century.”
The answer to this threat could be new antibiotics capable of conquering drug-resistant bacteria. Such pharmaceuticals most likely exist in nature, Brady says, because microbes competing for resources are in a constant tug-of-war. “The same drug-resistance mechanisms we see in hospitals are being used by soil bacteria trying to dodge their natural enemies,” he says, “and those enemies will in turn come up with new antibacterial substances.”
Since darwin’s time, ecologists have ventured across continents to explore new realms of biological diversity. Generations of microbiologists have trod this path as well.
After Fleming discovered the first antibiotic without leaving his lab in a London hospital, for example, other medical agents emerged from remote sites. The antibiotic erythromycin was obtained from bacteria residing in soil from the Philippines. Rapamycin, an immunosuppressant used to prevent the rejection of organ transplants, originated from the earth of Rapa Nui, more commonly known as Easter Island. And epothilone, a widely used anticancer agent, arose on the banks of South Africa’s Zambesi River.
“The field was based on the idea you would put on your explorer outfit and go hike the world looking for samples,” says Brady, whose lab has faithfully scrutinized dirt samples procured by volunteers from the deserts of Arizona, the Brazilian rainforest, and beyond.
Lately, however, he’s been sticking closer to home.
With help from the nonprofit Natural Areas Conservancy, Brady’s team has collected close to 300 soil samples from a variety of ecosystems within New York City parks, none more than 30 miles from the lab.
And, somewhat surprisingly, it turns out that they’ve got plenty to work with right here: New York’s topsoil is as cosmopolitan as the city that rests upon it. Among the DNA segments the researchers have extricated from park dirt, many look eerily familiar. Brady and his lab members have seen genes similar to those encoding erythromycin, rapamycin, and epothilone among other existing medicines, all of which were originally derived from microbes found elsewhere on the globe.
This suggests, according to the researchers, that the microbes from which many of our workhorse drugs were originally derived are not unique to their native environments. For example, Sorangium cellulosum, the African-born microbial architect of epothilone, may well have cousins and uncles in New York.

It’s excellent news for microbiologists with dwindling travel budgets or concerns about disturbing the world’s vanishing ecosystems.
“The fact that we can turn up the same potentially useful molecules in our own backyards suggests it may be time to shift away from a voyager approach,” Brady says, “and focus instead on the incredible abundance of unfamiliar genes that can be found right here.”
And indeed, despite the geographic commonalities, there’s a tremendous amount of unexplored chemistry among Brady’s samples. Of the millions of sequences the scientists have produced from New York City park dirt, less than one percent have matched with known genes in public databases. Lurking among the rest could be the next revolutionary pharmaceutical.
“There’s a tremendous amount we don’t recognize,” says Charlop-Powers. “A lot of new and potentially very useful chemistry is still out there.”